COVID-19 poses a riddle for the immune system
A dysregulated immune response, a cytokine storm and cytokine-release syndrome1,2 are some of the terms used to describe the overexuberant defence response that is thought to contribute to disease severity in certain people who become seriously ill with COVID-19. However, a precise definition of this type of immune dysfunction remains elusive. Writing in Nature, Lucas et al.3 fill in some gaps in our knowledge.
A holy grail of COVID-19 research is the ability to assess a person’s immune response, to pinpoint early the individuals who have mild symptoms but who are on track to develop the intense defence response that is associated with severe disease. This is important because there is a broad spectrum of clinical disease in people infected with SARS-CoV-2, the coronavirus that causes COVID-19: some infected individuals can be asymptomatic, whereas others are at risk of dying, and require hospitalization in an intensive-care unit and use of a ventilator machine to breathe4,5. Identifying those whose dysregulated immune-response signature predicts the development of severe disease would enable them to be monitored more intensively to minimize disease progression.
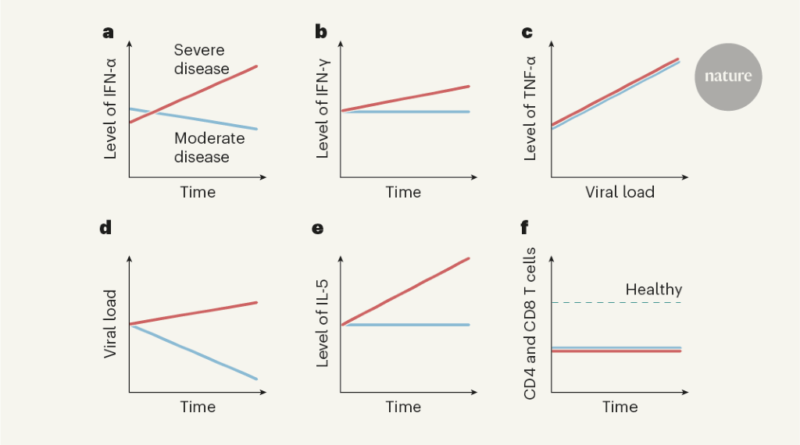
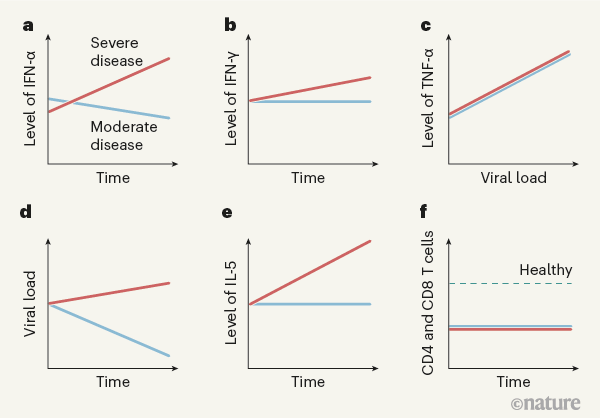
Lucas and colleagues performed extensive analyses of immune responses over time (longitudinal studies) in 113 people hospitalized with COVID-19 who had moderate or severe disease, and assessed a similar number of SARS-CoV-2-free healthy people as controls. The authors analysed molecules in blood plasma (Fig. 1) and monitored peripheral blood mononuclear cells — white blood cells of the immune system such as CD4 T cells, CD8 T cells and B cells. The longitudinal nature of this study enables conclusions to be drawn that wouldn’t be possible from analysing cross-sectional studies that don’t follow individuals over time.
The authors found that levels of several molecules that promote inflammation — immunomodulatory molecules termed cytokines, including IL-1α, IL-1β, IFN-α, IL-17A and IL-12 p70 — were higher in all of the people who had COVID-19 than in the healthy controls, providing a ‘core’ COVID-19 signature. Other cytokines, such as IFN-λ, thrombopoietin (which is associated with abnormalities in blood clotting), IL-21, IL-23 and IL-33, were upregulated to a greater extent in people with severe COVID-19 than in those with moderate disease. Several of the molecules upregulated in the core COVID-19 signature, as well as those seen in severe disease, have been identified previously as positively correlated with COVID-19 severity6,7. Severe disease was characterized by prolonged elevation of many of these molecules, whereas the levels of most of them subsided in people with moderate disease. Moreover, individuals with severe disease showed increased levels of cytokines associated with activation of a protein complex called the inflammasome, a component of the immune response that is a driver of inflammation. Also increased were levels of IL-1Ra, a protein that normally inhibits excessive inflammasome function, providing a rare example of an upregulated molecule that dampens the immune response in severe disease.
Levels of molecules associated with a defence response to viral infection — released by a type of activated CD4 T cell called a TH1 cell — were higher in people with severe disease than in those with moderate COVID-19. This occurred even though blood levels of CD4 T cells and CD8 T cells, which are generally linked to expression of these molecules, were similarly decreased (a condition called lymphopenia) in people with moderate or severe disease. More remarkably, cytokines associated with immune responses to fungi (cytokines released by a type of CD4 T cell called a TH17 cell) were elevated and remained so in people with severe disease. The same was true for cytokines associated with immune responses to parasites, including worms, or with allergic reactions (cytokines such as IL-5, released by a type of CD4 T cell called a TH2 cell). The discovery that parts of the immune system unrelated to viral control would be triggered by a viral infection was unexpected. Less surprising was the finding that levels of inflammatory cytokines in the blood, especially the proteins IFN-α, IFN-γ, TNF-α and TRAIL, correlated with viral RNA levels in the nasal passage, independently of disease severity.
From their analysis of proteins in people’s peripheral blood mononuclear cells, the authors divided individuals into three groups on the basis of their subsequent clinical course and disease severity. In general, at early time points after infection, those who went on to have moderate disease had low levels of inflammatory markers and a rise in the level of proteins associated with tissue repair. By contrast, people who went on to develop severe or very severe disease had increased expression of IFN-α, IL-1Ra and proteins associated with TH1-, TH2- and TH17-cell responses, even at early time points (10–15 days after the onset of symptoms). These results were validated using data for the entire patient population, across all time points, thus demonstrating that these characteristic expression patterns persisted over time in people with each type of disease severity.
What have we learnt from this report, and what still needs to be done? It is clear from this and other studies that the immune response in hospitalized patients with severe COVID-19 is characterized by lymphopenia and the expression of molecules associated with ongoing inflammation8, whereas these same molecules are expressed at a lower level in people with mild or moderate disease. Differences in immune responses between the different categories of disease severity are even more evident when people with very mild or subclinical disease are included in the analyses4.
A key next step will be to analyse samples from people with extremely early signs of COVID-19, and to compare longitudinal data in those who do and those who don’t require hospitalization. Some people who develop severe disease seem to have a suboptimal immune response initially, which might allow uncontrolled viral replication9. Such high replication might, in turn, contribute to severe disease.
Further analyses should identify molecules that are useful for predicting which individuals will later be hospitalized and require intensive care. It will also be crucial to understand how severe disease results in an upregulation of cytokines usually linked to the immune response to parasites and allergic reactions, and whether this apparent dysregulation of the immune response to viral infection is unique to COVID-19. It will also be worth determining whether these changes in the expression of inflammatory molecules in the blood also occur in cells at the site of infection — the airways and lungs. Lucas et al. analysed blood samples because obtaining cells from an infected lung is much more tricky and results in the production of aerosols that might contain SARS-CoV-2.
For results to be clinically useful, it will be necessary to define a limited number of biomarkers that can be both readily measured and used to predict disease outcomes. This could be difficult, because many of the changes in cytokine expression observed in studies such as that of Lucas and colleagues are useful for population-level analyses but less so for predicting outcomes in individual patients. Levels of specific cytokines vary substantially between people, making it hard to benchmark a level of cytokine expression that constitutes a sign of abnormality. Therefore, groups of cytokines, each with different degrees of inter-individual variability, must be measured to identify useful alterations.
The identification of infected people on course to develop severe COVID-19 will be a key step forward in patient care. For example, it would increase the possibility of correctly selecting individuals most in need of targeted early treatment, such as with therapies that directly inhibit viral replication. There has been progress in identifying such treatments, and the continued development of antiviral drugs that have increased efficacy and specificity will be crucial for alleviating the disease and reducing the death rate associated with the COVID-19 pandemic. Ideally, such drugs will be administered orally, and will reduce the need for hospitalization. Continued progress in unravelling the immune response to SARS-CoV-2 infection will help to improve clinical treatments for COVID-19.